Mating
mate(cohort_A ::Cohort,
cohort_B ::Cohort;
nA ::Int64=cohort_A.n,
nB_per_A ::Int64=1,
n_per_mate ::Int64=1,
replace_A ::Bool =false,
replace_B ::Bool =false,
ratio_malefemale ::Union{Float64, Int64}=0,
scheme ::String ="none",
args...)
mate(cohort::Cohort; args...) = mate(cohort, cohort; args...)Arguments
Positional arguments
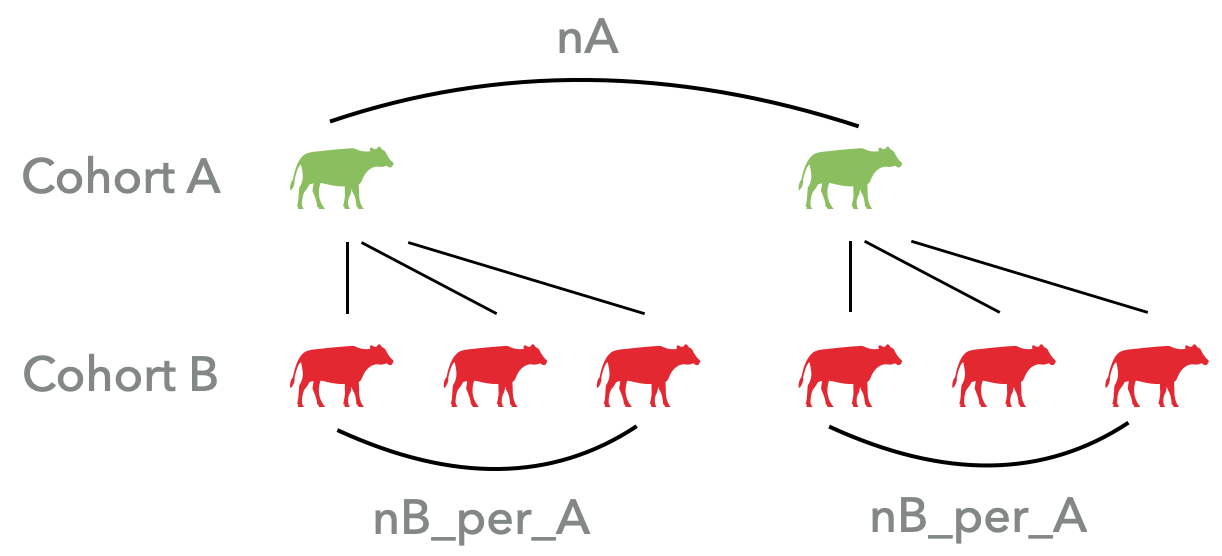
cohort_A: Acohortobject that is treated as common mating parents.cohort_B: Acohortobject that is a mating pool from which individuals are sampled to mate withcohort_A.
Keyword arguments
nA:nAindividuals will be sampled fromcohort_Aand treated as common parents.nB_per_A:nB_per_Aindividuals sampled fromcohort_Bwill mate with each individual fromcohort_A.n_per_mate:n_per_mateprogenies will be reproduced from each pair of mating parent.replace_A: Whether the sampling is replacable incohort_A.replace_B: Whether the sampling is replacable incohort_B.ratio_malefemale: By default, two cohorts which are male and female progenies will be returned.ratio_malefemaledefined the progenies ratio of males over females. Ifratio_malefemale=0, only one cohort will be returned.scheme: Available options are ["random", "diallel cross", "selfing", "DH"]. See the examples for more details.
Outputs
By default, two cohort objects will be returned. The first cohort is assumed to be male progenies and the other cohort are female progenies. The size of two cohorts will folow the ratio raiot_malefemale. When ratio_malefemale is set to 0, only one cohort will be returned.
Examples
Random Mating (Default)
Initialize cohorts
julia> cohort_A = Cohort(5)
julia> cohort_B = Cohort(10)Define mating events
julia> args = Dict(:nA => cohort_A.n,
:nB_per_A => 1,
:replace_A => false,
:replace_B => false,
:n_per_mate => 1)
julia> progenies = mate(cohort_A, cohort_B; args...)
# Equivalent
julia> progenies = mate(cohort_A, cohort_B)
# Equivalent
julia> progenies = mate(cohort_A, cohort_B; scheme="random")
# Equivalent
julia> progenies = cohort_A * cohort_BCheck the pedigree to see if the mating goes as desired.
julia> get_pedigree(progenies)
5×3 LinearAlgebra.Adjoint{Int64,Array{Int64,2}}:
19 1 8
16 2 6
17 3 10
20 4 15
18 5 14Diallel Cross
Initialize cohorts
julia> cohort_A = Cohort(2)
julia> cohort_B = Cohort(5)Define mating events
julia> args = Dict(:nA => sires.n,
:nB_per_A => dams.n,
:replace_A => false,
:replace_B => false,
:n_per_mate => 1,
:ratio_malefemale=> 1)
julia> male, female = mate(cohort_A, cohort_B; args...)
# Equivalent
julia> male, female = mate(cohort_A, cohort_B; scheme="diallel cross")Check the pedigree to see if the mating goes as desired.
julia> get_pedigree(male + female)
10×3 LinearAlgebra.Adjoint{Int64,Array{Int64,2}}:
12 2 7
10 2 6
11 2 4
14 1 3
15 1 5
9 2 5
13 1 6
17 1 4
8 2 3
16 1 7Selfing
Initialize cohorts
julia> parents = Cohort(5)In the selfing scheme, only one cohort is required.
julia> args = Dict(:nA => 3,
:replace_A => false,
:n_per_mate => 5,
:scheme => "selfing")
julia> progenies = mate(parents; args...)Inspect the pedigree to verify the mating behavior
julia> get_pedigree(progenies)
15×3 LinearAlgebra.Adjoint{Int64,Array{Int64,2}}:
6 4 4
7 4 4
8 4 4
9 4 4
10 4 4
11 1 1
12 1 1
13 1 1
14 1 1
15 1 1
16 5 5
17 5 5
18 5 5
19 5 5
20 5 5Mating with the pedigree defined
Define a genome containing 4 loci
julia> build_genome(n_loci=4, n_chr=1)Simulate a trait controlled by 2 loci
julia> build_phenome(2, h2=.5)Load demo pedigree data frame
julia> ped = DATA("pedigree")
5×3 DataFrame
Row │ Column1 Column2 Column3
│ Int64 Int64 Int64
─────┼───────────────────────────
1 │ 1 0 0
2 │ 2 0 0
3 │ 3 0 0
4 │ 4 1 2
5 │ 5 1 2Create a cohort
# Option 1: with simulated genotypes
julia> cohort = Cohort(3)
# Option 2: with real genotypes
julia> genotypes = DATA("genotypes")
5×4 DataFrame
Row │ Column1 Column2 Column3 Column4
│ Int64 Int64 Int64 Int64
─────┼────────────────────────────────────
1 │ 2 0 0 1
2 │ 0 0 1 0
3 │ 0 1 0 2
4 │ 1 1 0 2
5 │ 2 0 2 0
julia> cohort = Cohort(genotypes)
julia> cohort|>get_genotypes
5×4 Array{Int64,2}:
2 0 0 1
2 0 2 0
0 0 1 0
0 1 0 2
1 1 0 2Generate offsprings with the defiend pedigree
julia> offsprings = mate(ped)Examine the genotypes
julia> offsprings |> get_genotypes
5×4 Array{Int64,2}:
2 0 0 1
2 0 2 0
0 0 1 0
2 0 1 0
2 0 1 0